Part:BBa_K4292021
X-3-gmd-wcaG
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 5089
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 5346
Illegal BsaI.rc site found at 5506
Contribution
Our team aims to produce 2'-FL in S. cerevisiae for providing the economic strategy. In terms of raw material, there are two pathways to produce GDP-L-fucose from GDP-D-mannose (called de novo pathway) or L-fucose (called salvage pathway). In the de novo pathway, the GDP-D-mannose converts 2'-FL and secreted into medium involving three enzymes gmd, wcaG, and wbgL, as well as transporter lac12. Our team aims to produce 2'-FL in S. cerevisiae for providing the economic strategy. In terms of raw material, there are two pathways to produce GDP-L-fucose from GDP-D-mannose (called de novo pathway) or L-fucose (called salvage pathway). In the de novo pathway, the GDP-D-mannose converts 2'-FL and secreted into medium involving three enzymes gmd, wcaG, and wbgL, as well as transporter lac12.
In order to obtain the high yield of 2'-FL, we introduced the four exogenous gene including gmd, wcaG into S. cerevisiae genome using CRISPER-cas9. As shown in Figure1. The genes, gmd, wcaG , are encoding the important enzymes involving the 2'-FL synthesis. gmd and wcaG are converted the abundant intracellular GDP-mannose in S. cerevisiae into GDP-fucose. Further, 2'-FL can be produced from GDP-fucose by expression of Wbgl and metabolization. In this work, we used sweet potato residues as main carbon source.
Engineering Success
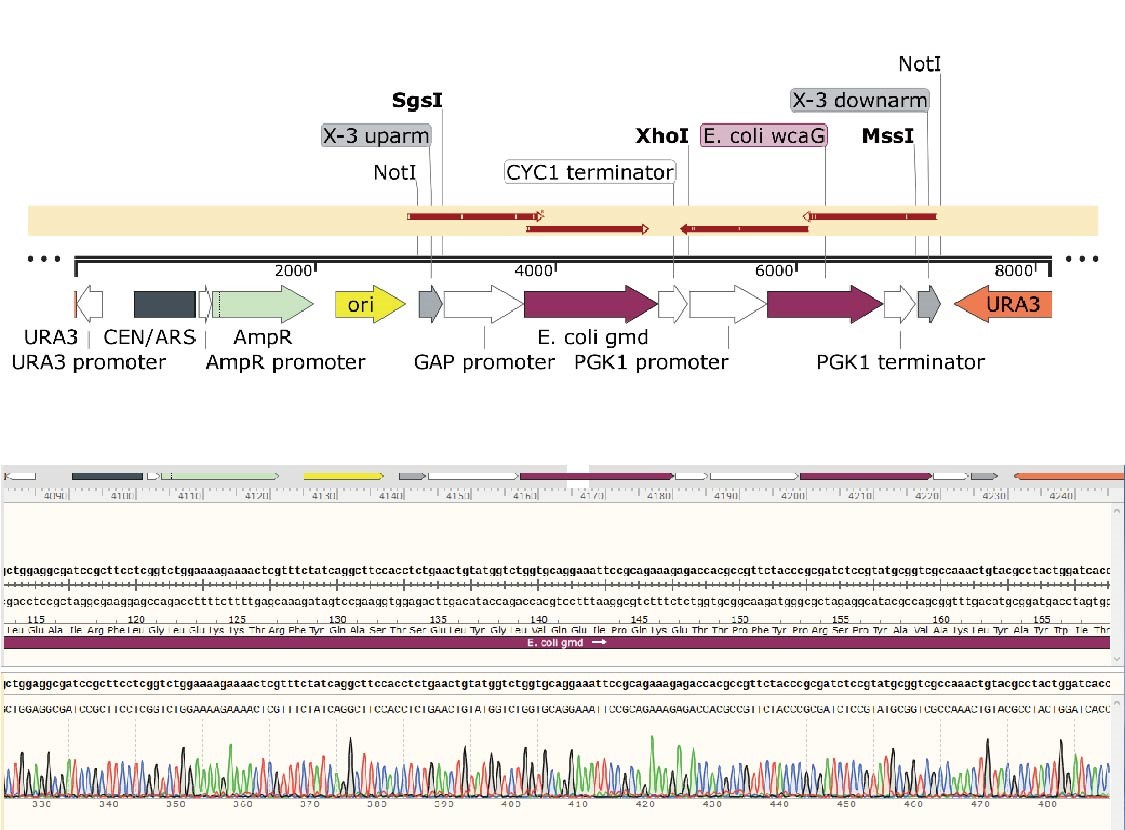
We constructed two plasmids including two genes, gmd and wacG which are key genes to produce 2'-Fucosyllactose(2'-FL) in a yeast cellular factory. In addition, we need to added the promoter and terminator to flanking regions of these exogenous genes in order to facilitate expression in the engineered yeast. The components were incorporated into the integration backbone plasmid with NotI and XhoI sites. Firstly, we constructed the integration backbone plasmids XI-2 and X-3. These colonies were verified by colony PCR and Sanger sequencing and the results were shown as follows.
In the X-3-gmd-wcaG plasmid, the gmd and wcaG gene expression cassettes are inserted between the upstream and downstream of the X-3 homology arm, in the same direction. The gmd+wcaG gene for X-3 site integration was constructed by a two-step digestion cloning method. After the gmd gene expression cassette was integrated, the wcaG was introduced using Xho1 digestion site.
Plasmids of 12 transformants were extracted and verified by Xho1+Mss1 double-enzyme digestion (Figure 3). The positive transformant band was 6220+1896 bp, and the No.11 plasmid with correct digestion was randomly selected and sent for sequencing.
The details of sequence alignment showed there is no mutation, indicated that the X-3-gmd-wcaG plasmid was constructed successfully.
| None |